Information about course databases
clinical_vocabulary
This is essentially an OMOP database without PHI, just vocabularies.
I downloaded a number of vocabularies from AthenaSpecifically, I pulled CDM 5 vocabularies having the following vocabulary codes: SNOMED, ICD9CM, ICD9Proc, CPT4, HCPCS, LOINC, NDFRT, RxNorm, NDC, Gender, Race, MedDRA, Read, ATC, VA Product, VA Class, Cohort, ICD10, Ethnicity, MeSH, ICD10CM, RxNorm , Nebraska Lexicon, OMOP Extension. and just inserted them into new tables. I added a number of helpful indices, which dramatically improve the performance.
This is the main database you’ll be using.
drugbank
I downloaded the full DrugBank XML database dump, version 5.1.7.
I used dbparser version 1.2.0 to automatically reformat this XML document into tables.
Most of these tables were inserted directly into the database, though a few smaller, less-important tables were omitted.
You’re most likely to use this database to access natural language drug indicationsdrugs_pharmacology.indication
, though you’re free to use the wealth of other information available.
SIDER
Tables were downloaded from SIDER, version 4.1.
I did not reorder columns, but I did define names where none were present.
See the SIDER 4.1 README for more information about the columns in these tables.
In particular, note that drug names are automatically generated in the drug_names table.
You may want to use names from ATC codes instead (i.e. map SIDER.drug_atc.drug_atc_code to clinical_vocabulary.CONCEPT.concept_code to access the concept_name field.)
faers
This contains some data from FDA adverse event reports from the second quarter of 2020. I renamed only tables, not fields. For documentation, see the ASC_NTS.pdf.
Favorite online resources
- Athena
- ↑ The most comprehensive front-end for OMOP vocabulary tables of which I’m aware. Functional interface. ↓ Can be a bit slow, and mappings aren’t named in an obvious way. Can’t collect codes like in Atlas.
- I use this on an almost daily basis.
- Book of OHDSI
- This thing is insanely good. It has information about OMOP, which is most relevant for this class. That’s only a small fraction, though. It has detailed explanations of OHDSI tools, observational research methods, and all kinds of other things. Plus, you’ll recognize many of the authors from our department.
- Lab tests online
- Amazing resource for lab tests. Great place to look for reference ranges, LOINC codes, related tests, etc.
- ICD10Data.com
- ↑ Adds helpful clinical information for ICD-10 codes. Very Google-able. Comprehensive for ICD-10. ↓ Internal search isn’t always great. Not the best interface for finding relatives of a code.
- DrugBank
- Contains tons of information about drugs, including indications, biology/chemistry/physics data, cross-links to other data sources, etc.
- Tends to be my go-to for looking at a new drug.
- OBO Foundry
- Huge resource of open-source, standardized, interoperable biomedical ontologies. Includes well known resources like the Gene Ontology, ChEBI, etc.
Personal software choices
Throughout this course, you are free to use whatever setup you prefer. It makes no difference to me, and I want you to use what you find most comfortable.
Sometimes, though, I like to see what other people use.
For SQL, I use DataGrip.
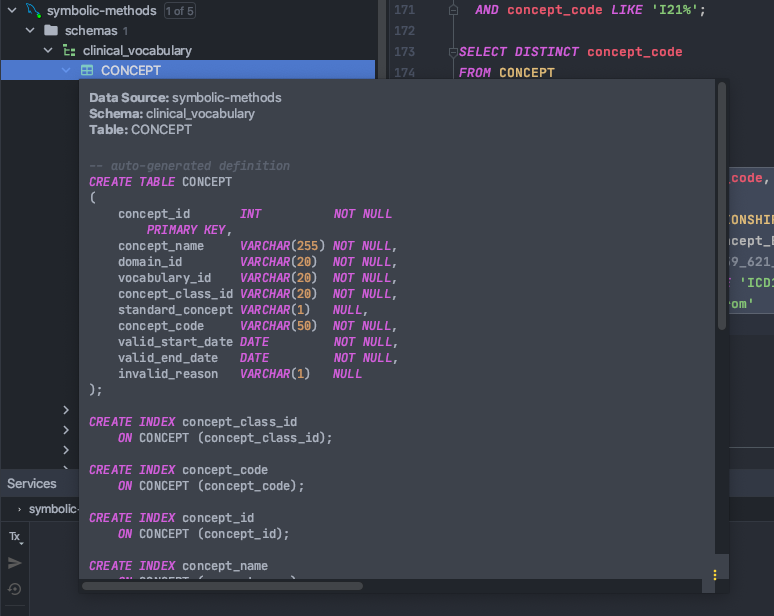
Example auto-definition
It has a great built-in editor.
I can very easily run queries in parallel or in a queue.
It does nice auto code reformatting, which keeps you SQL files looking clean.
It also stores the database schema and can generate table definitions, which made creating the class database insanely easy.
You can get DataGrip free with a student account.
For statistical analysis, I prefer R + tidyverse. For slightly more complex code, I like Python (typically install miniconda). For both, I’m a big fan of Jupyter notebooksIRKernel for R and conda.
I’d also highly recommend Manubot, which enables automated scholarly manuscripts on GitHub.